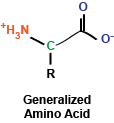As mentioned in the last post, several types of cells derive from lymphoid progenitor cells. These cells are generated in the bone marrow in general, but only B cells mature there (hence the name B cells). In contrast, T cells migrate to the thymus where they mature. After full maturation of both B and T cells, they circulate in the blood system and then enter the peripheral lymphoid organs. The central lymphoid organs are the bone marrow where the lymphocytes are generated, whereas the peripheral lymphoid organs are where T cells mature and where the adaptive immune responds to various stimuli.
The peripheral lymphoid organs
First, we will consider the components of the peripheral lymphoid system. The lymph nodes are glands right near each armpit that is where fluid collects from the lymph system. Lymph drains into the lymph nodes via lymphatic vessels and consists of the extracellular fluid filtered from the blood. Thus, lymph is kind of a surveillance system for the body. The afferent lymphatic vessels carry lymph and cells from infected tissues and drain into the lymph node.
The lymph node itself has a unique structure, illustrated to the right. The follicles are where B lymphocytes set up shop, and T cells exist in paracortical areas (T-cell zones). Germinal centers within the lymph node are where B cells proliferate after they have been stimulated by T cells. Several additional tissues are organized similar to the lymph node drawn to the right, and this structure facilitates interaction between B and T cells.
The spleen is another peripheral lymphoid organ that mostly works to break down dead red blood cells. This destruction occurs in the red pulp of the spleen, but the spleen also has white pulp where lymphocytes enter and exist within the spleen. Within the white pulp is the periarteriolar lymphoid sheath (PALS) that contains T cells and a B-cell corona.
The digestive system is a major route for infection and has several gut-associated lymphoid tissues (GALT). Some of these tissues include the tonsils, adenoids, and the appendix. The intestine also has its own GALT, namely the Peyer's patches, which collect antigen directly from inside the intestine using multi-fenestrated (M) cells.
Similar to the digestive tract, the respiratory tract has its own lymphoid tissue, called the bronchial-associated lymphoid tissue (BALT).
Monday, November 14, 2011
Wednesday, November 9, 2011
The basics of immunology
Immunology scares me. I'm not ashamed to admit this fact. I find the topic intimidating and overwhelming, especially when I listen to talks given by prominent immunologists. The terminology is difficult, and the concepts seem very intertwined. I've always perceived that breaking into understanding immunology required a lot of work but that it would (and should) make sense... eventually.
The next few blog posts are going to focus on immunology, not only because I need to learn this information, but also because it is fascinating and a challenging topic.
Components of the Immune System
All of the cells that comprise the immune system emerge from the bone marrow, where all of them originally come from and where some of them remain for maturation. The cell type that gives rise to immune cells is the hematopoietic stem cell. From this pluripotent state, the hematopoetic stem cell can then mature into a myeloid progenitor cell or a common lymphoid progenitor. Myeloid progenitor cells can differentiate into several more cell types, including granulocyte and macrophage progenitors and megakaryocyte and erythrocyte progenitors. The granulocyte and macrophage progenitors can then develop into neutrophils, eosinophils, basophils, mast cells, and macrophages. Megakaryocyte and erythrocyte progenitors generate platelets upon maturation.
Hematopoietic stem cells can also develop into a common lymphoid progenitor, which consists of B cells, T cells, and NK cells. These types of cells leave the bone marrow and migrate through the lymph nodes. Dendritic cells also develop from lymphoid progenitor cells but mature in the bone marrow before entering the lymph node.
Basic functions of immune cells
The next few blog posts are going to focus on immunology, not only because I need to learn this information, but also because it is fascinating and a challenging topic.
Components of the Immune System
All of the cells that comprise the immune system emerge from the bone marrow, where all of them originally come from and where some of them remain for maturation. The cell type that gives rise to immune cells is the hematopoietic stem cell. From this pluripotent state, the hematopoetic stem cell can then mature into a myeloid progenitor cell or a common lymphoid progenitor. Myeloid progenitor cells can differentiate into several more cell types, including granulocyte and macrophage progenitors and megakaryocyte and erythrocyte progenitors. The granulocyte and macrophage progenitors can then develop into neutrophils, eosinophils, basophils, mast cells, and macrophages. Megakaryocyte and erythrocyte progenitors generate platelets upon maturation.
Hematopoietic stem cells can also develop into a common lymphoid progenitor, which consists of B cells, T cells, and NK cells. These types of cells leave the bone marrow and migrate through the lymph nodes. Dendritic cells also develop from lymphoid progenitor cells but mature in the bone marrow before entering the lymph node.
Basic functions of immune cells
- Macrophages are a common cell type that mature from monocytes (from the myeloid progenitor cells originally). Monocytes circulate in the blood and continuously differentiate into macrophages when they enter the body's tissues. Once in the tissues, macrophages can be considered the garge trucks of the body: they engulf the environment as well as other cells in the process of phagocytosis. Thus, macrophages can function to neutralize harmful elements within the body.
- Dendritic cells also mature from myeloid progenitor cells, and their main function is to process and display antigen that will then be readable by T lymphocytes. This antigen display requires the presentation of co-stimulatory molecules, and when dendritic cells encounter a pathogen (or other foreign antigen), they mature and begin expressing these co-stimulatory molecules.
- Mast cells differentiate in body tissues and are involved in mediating mucosal immunity. They are most well-known for their role in allergic reactions.
- Neutrophils are a type of granulocyte (so called because they have densely-staining and strange-shaped nuclei) that are involved in phagocytosis and increase in numbers upon an immune response.
- Eosinophils respond to parasites.
- Basophils may function similarly to mast cells.
- B cells differentiate into plasma cells and function to secrete antibodies.
- T cells destroy virus-infected cells and also function to activate other immune cells, such as B cells and macrophages.
- NK cells are involved in innate immunity and destroy "weird-looking" cells, such as tumor cells or cells infected with viruses.

References for the interested:
Immunobiology. Janeway, Travers, Walport, Shlomchik.
Basic Concepts of Immunology and Neuroimmunology: Basic Immunology
Basic Concepts of Immunology and Neuroimmunology: Basic Immunology
Wednesday, November 2, 2011
Cancer and Oncogenes
Cancer is a diverse group of diseases with one common characteristic: unchecked cellular replication. Via several potential mechanisms, cancer cells are able to avoid all of the checkpoints involved in cell growth and division, thus enabling them to divide more frequently or indefinitely. Many events can lead to the development of a cancer cell, including inheritance of mutated DNA or the activity of a carcinogen, or a chemical agent that leads to the development of cancer.
Gene expression is often deregulated in cancer cells such that some genes are overexpressed, while others are underexpressed. Genes that can be mutated to lead to an upregulation of activity and lead to the development of a cancer cell are termed proto-oncogenes. When these proto-oncogenes are actually mutated, they are considered oncogenes.
 An oncogene is often a gene involved in regulating cell division and drive the cell cycle. When they are overexpressed, such as during cancer, they can push the cell to divide more frequently and, with further mutations, transform the cell such that it divides without restriction.
An oncogene is often a gene involved in regulating cell division and drive the cell cycle. When they are overexpressed, such as during cancer, they can push the cell to divide more frequently and, with further mutations, transform the cell such that it divides without restriction.
Oncogenes were first discovered in viruses, specifically in Rous Sarcoma Virus (RSV), a retrovirus that encodes a homologue to cellular src kinase (the viral form called v-src). Tumors in birds caused by RSV are the result of v-src causing unregulated cellular proliferation. Large amounts of research into this area has identified cellular src kinase as a proto-oncogene that, when mutated to become constitutively active, becomes an oncogene and can drive cancer development. Interestingly, viruses have highlighted a number of cellular oncogenes and pathways that are improperly regulated in cancer. Over 20 viral oncogenes have been identified to date.
Cellular proto-oncogenes (the genes before they become oncogenes) can promote cellular proliferation and the development of cancer in several ways. One of these ways is to be overexpressed and function when the gene product really shouldn't function. This is the case with proteins such as myc and growth factor receptors. With overexpression of these proteins, there is the potential for amplified signaling through these pathways that can push the cell to divide more than it normally does, leading to the development of cancer. An additional mechanism whereby a proto-oncogene can become an oncogene is via mutation that leads to improper regulation, such as constitutive activity. A classical example of this type of phenomenon is via Ras, which when mutated is constitutively active and cannot hydrolyze an attached GTP to inactivate. Thus, Ras remains active and cannot be "turned off." This constant activity of Ras results in signal transduction to the nucleus of the cell and pushes the cell to divide through transcription of several genes involved in cell division.
Gene expression is often deregulated in cancer cells such that some genes are overexpressed, while others are underexpressed. Genes that can be mutated to lead to an upregulation of activity and lead to the development of a cancer cell are termed proto-oncogenes. When these proto-oncogenes are actually mutated, they are considered oncogenes.
 An oncogene is often a gene involved in regulating cell division and drive the cell cycle. When they are overexpressed, such as during cancer, they can push the cell to divide more frequently and, with further mutations, transform the cell such that it divides without restriction.
An oncogene is often a gene involved in regulating cell division and drive the cell cycle. When they are overexpressed, such as during cancer, they can push the cell to divide more frequently and, with further mutations, transform the cell such that it divides without restriction. Oncogenes were first discovered in viruses, specifically in Rous Sarcoma Virus (RSV), a retrovirus that encodes a homologue to cellular src kinase (the viral form called v-src). Tumors in birds caused by RSV are the result of v-src causing unregulated cellular proliferation. Large amounts of research into this area has identified cellular src kinase as a proto-oncogene that, when mutated to become constitutively active, becomes an oncogene and can drive cancer development. Interestingly, viruses have highlighted a number of cellular oncogenes and pathways that are improperly regulated in cancer. Over 20 viral oncogenes have been identified to date.
Cellular proto-oncogenes (the genes before they become oncogenes) can promote cellular proliferation and the development of cancer in several ways. One of these ways is to be overexpressed and function when the gene product really shouldn't function. This is the case with proteins such as myc and growth factor receptors. With overexpression of these proteins, there is the potential for amplified signaling through these pathways that can push the cell to divide more than it normally does, leading to the development of cancer. An additional mechanism whereby a proto-oncogene can become an oncogene is via mutation that leads to improper regulation, such as constitutive activity. A classical example of this type of phenomenon is via Ras, which when mutated is constitutively active and cannot hydrolyze an attached GTP to inactivate. Thus, Ras remains active and cannot be "turned off." This constant activity of Ras results in signal transduction to the nucleus of the cell and pushes the cell to divide through transcription of several genes involved in cell division.
Labels:
cancer,
cell cycle,
oncogene,
oncogenesis,
proto-oncogene,
ras,
rous sarcoma virus
Monday, October 17, 2011
The Trp Operon
The last post about an operon (the lac operon) is the most viewed post on this blog, so I thought that it might be helpful to follow this up with another operon, this time concentrating on the trp (tryptophan) operon. This operon is another really elegant example of transcriptional regulation in E.coli and the mechanism is pretty cool.
Amino acids are essential for life (see the last post on their composition!) and cells synthesize amino acids using a variety of enzymes. When nutrients are plentiful, such as E.coli would encounter in nutrient broth in the laboratory setting, cells no longer need to waste energy producing biosynthetic enzymes when they can utilize nutrients already in excess. The trp operon contains several enzymes that are coordinately regulated and involved in the production of tryptophan. When tryptophan is present in the cell's environment, it doesn't need to make any of these enzymes, but if the cell needs tryptophan, these enzymes are transcribed and shortly thereafter translated. Control of this operon, thus, controls how much energy the cell is going to put into making tryptophan.
Similar to the lac operon, the trp operon contains an operator (O) sequence, within the promoter sequence, where an operator binds and prevents transcription. In the presence of tryptophan, the operator binds the promoter and prevents RNA polymerase from transcribing genes. In the absence of tryptophan, however, transcription occurs at a basal rate. Sounds simple enough, right? Let's take it a step further and consider...
Attenuation
An important concept in gene regulation is that of attenuation, which is fine-tuning of gene expression. You might think that attenuation is mediated by protein factors that bind the DNA and affect gene expression; however, attenuation of the trp operon is a little different and, instead, depends on mRNA structure to modulate gene expression.
Before moving forward, let's look at the trp operon (diagrammed to the right). Briefly,t here are four regions, and these four regions have differing levels of complementarity to each other. Thus, when the DNA is transcribed into mRNA, the mRNA folds into all kinds of shapes and the regions of the trp operon fold on each other.
After transcription of the entire trp operon (we're dealing with mRNA from this point forward), the next event is translation of this mRNA into protein. In bacteria, it's important to note that transcription and translation occur simultaneously, so as soon as we have a transcript in a bacterial cell, it's being translated. The trp transcript contains two critical tryptophan codons immediately before region 1, so in order to synthesize the enzymatic machinery to make tryptophan, the cell must use a few residues to translate the protein.
In the presence of high amounts of tryptophan within the cell, the ribosome plows through these two tryptophan codons, adding in the appropriate amino acids, and continuing through region 1 of the mRNA. This results in region 1 and 2 mRNA sequences binding together, and then regions 3 and 4 bind together as well. This interaction between regions 3 and 4 results in the creation of a transcription-termination hairpin, basically a structure in the mRNA that kicks out RNA polymerase and prevents further transcription of the mRNA. Thus, transcription (and then translation) are stopped because
In the absence of tryptophan, however, the ribosome cannot quickly add tryptophan during the translation process and it stalls before region 1. This results in the folding of the mRNA such that regions 2 and 3 bind to each other. When this structure forms, no transcriptional termination hairpin is formed, and mRNA synthesis continues. Thus, the entire mRNA sequence for the trp operon is made and can be translated into enzymes that will synthesize tryptophan.
In summary:
Lots of tryptophan: Ribosome zooms through the mRNA, regions 1 & 2 and 3 & 4 bind (in pairs) and create a termination hairpin
End result: Transcription terminates and tryptophan synthetic enzymes not created (cell saves energy!)
Lack of tryptophan: Ribosome stalls immediately before region 1, regions 2 and 3 bind each other, no termination hairpin is formed
End result: Transcription continues and biosynthetic enzymes are eventually synthesized
This scheme is similar for other operons encoding amino acid biosynthetic enzymes (in bacteria, that is). The trp operon is an elegant scheme to finely-tune transcription via mRNA structure to prevent the cell from wasting energy.
Amino acids are essential for life (see the last post on their composition!) and cells synthesize amino acids using a variety of enzymes. When nutrients are plentiful, such as E.coli would encounter in nutrient broth in the laboratory setting, cells no longer need to waste energy producing biosynthetic enzymes when they can utilize nutrients already in excess. The trp operon contains several enzymes that are coordinately regulated and involved in the production of tryptophan. When tryptophan is present in the cell's environment, it doesn't need to make any of these enzymes, but if the cell needs tryptophan, these enzymes are transcribed and shortly thereafter translated. Control of this operon, thus, controls how much energy the cell is going to put into making tryptophan.
Similar to the lac operon, the trp operon contains an operator (O) sequence, within the promoter sequence, where an operator binds and prevents transcription. In the presence of tryptophan, the operator binds the promoter and prevents RNA polymerase from transcribing genes. In the absence of tryptophan, however, transcription occurs at a basal rate. Sounds simple enough, right? Let's take it a step further and consider...
Attenuation
An important concept in gene regulation is that of attenuation, which is fine-tuning of gene expression. You might think that attenuation is mediated by protein factors that bind the DNA and affect gene expression; however, attenuation of the trp operon is a little different and, instead, depends on mRNA structure to modulate gene expression.
Before moving forward, let's look at the trp operon (diagrammed to the right). Briefly,t here are four regions, and these four regions have differing levels of complementarity to each other. Thus, when the DNA is transcribed into mRNA, the mRNA folds into all kinds of shapes and the regions of the trp operon fold on each other.
After transcription of the entire trp operon (we're dealing with mRNA from this point forward), the next event is translation of this mRNA into protein. In bacteria, it's important to note that transcription and translation occur simultaneously, so as soon as we have a transcript in a bacterial cell, it's being translated. The trp transcript contains two critical tryptophan codons immediately before region 1, so in order to synthesize the enzymatic machinery to make tryptophan, the cell must use a few residues to translate the protein.
In the presence of high amounts of tryptophan within the cell, the ribosome plows through these two tryptophan codons, adding in the appropriate amino acids, and continuing through region 1 of the mRNA. This results in region 1 and 2 mRNA sequences binding together, and then regions 3 and 4 bind together as well. This interaction between regions 3 and 4 results in the creation of a transcription-termination hairpin, basically a structure in the mRNA that kicks out RNA polymerase and prevents further transcription of the mRNA. Thus, transcription (and then translation) are stopped because
In the absence of tryptophan, however, the ribosome cannot quickly add tryptophan during the translation process and it stalls before region 1. This results in the folding of the mRNA such that regions 2 and 3 bind to each other. When this structure forms, no transcriptional termination hairpin is formed, and mRNA synthesis continues. Thus, the entire mRNA sequence for the trp operon is made and can be translated into enzymes that will synthesize tryptophan.
In summary:
Lots of tryptophan: Ribosome zooms through the mRNA, regions 1 & 2 and 3 & 4 bind (in pairs) and create a termination hairpin
End result: Transcription terminates and tryptophan synthetic enzymes not created (cell saves energy!)
Lack of tryptophan: Ribosome stalls immediately before region 1, regions 2 and 3 bind each other, no termination hairpin is formed
End result: Transcription continues and biosynthetic enzymes are eventually synthesized
This scheme is similar for other operons encoding amino acid biosynthetic enzymes (in bacteria, that is). The trp operon is an elegant scheme to finely-tune transcription via mRNA structure to prevent the cell from wasting energy.
Labels:
amino acids,
gene regulation,
lac operon,
operator,
operon,
transcription,
trp,
tryptophan
Thursday, October 13, 2011
Amino Acids: The Building Blocks of Proteins
While I've written many posts describing signaling pathways and cellular phenomena of significant complexity, I'd like to use this post to take a step back and look at some fundamental building blocks, first turning to amino acids, the monomers that, when polymerized, make up polypeptides and proteins.
At the most basic level, amino acids are really a rather simple chemical compound, consisting of a amino group, a carboxy group, a hydrogen, and a side chain, all sticking off a central carbon atom. These amino acids are polymerized via their amino and carboxy chemical groups to create long, linear linkages.
Brief aside: In my chemistry class in undergrad, my TA helped us remember the order of the chemical bonds following polymerization by saying N-H, C-H, C-O, N-H, C-H, C-O, ...
You may remember briefly from any stint in chemistry class that a carbon atom that is covalently bound to four different chemical entities (in this case, a side chain, a hydrogen atom, a carboxyl group, and an amino group) can take two different conformations, depending on how these bonds are spatially oriented. In the case of amino acids, the vast majority of amino acids found in our bodies and used to generate proteins are L stereoisomers. This is a result of the amino acid synthesis machinery structure exclusively generating L amino acids. There are exceptions, but we won't get into that.
As I mentioned, amino acids have a side chain: the part of the amino acid that endows it with its identity. These side chains can be broken into a few groups that we will explore now:
The first set of side chains is the nonpolar, hydrophobic side chains. The amino acids in this group include alanine, valine, leucine, isoleucine, glycine, methionine, and proline (structures shown to the right). What you'll immediately notice is that these amino acid side chains are composed mostly of hydrogens and carbons. Thus, these side chains do not contain polar covalent bonds and do not interact as readily with water (thus the term hydrophobic - they're "afraid" of water. Some amino acids of note in this group are proline, which contains a ring structure that creates a "kink" in the amino acid, and methionine, which contains a sulfur atom.
The next set is composed of the aromatic side chains, which includes phenylalanine, tyrosine, and tryptophan. These amino acids all contain an aromatic ring, which makes them relatively nonpolar; thus, they do not interact favorably with water. These amino acids are involved in mediating protein protein interactions and are frequently found at the active sites of enzymes.
Next up: polar, uncharged side chains: asparagine, cysteine, glutamine, serine, and threonine. These amino acids contain hydroxyl, sulfhydryl, or amide groups that mediate interactions with water, but they carry no net charge. An amino acid of note in this family is cysteine, which can react with itself to form cystine, which is important in mediating the formation of disulfide bonds in protein structures.
We'll consider basic side chains next. These amino acids consist of arginine, histidine, and lysine, which all carry a net positive charge in solution. Of note, histidine is commonly found at the active site of enzymes to serve as a protein donor or acceptor.
Finally, we find acidic side chains: aspartate and glutamate. In solution, these amino acids carry a negative charge and are considered acidic.
In the diagram at right, I've drawn up each of the amino acids along with their three-letter and one-letter codes. These codes are frequently used to abbreviate long lists of amino acids.
Another brief aside: Did you know that a single woman designated the amino acid abbreviations? She chose letters that made sense for most amino acids (as you can see above). For tryptophan, for example, she chose W because she envisioned saying tryptophan as twyptophan. Kind of cool, huh?
As a summary, here are the amino acid abbreviations:
So there you have it: 20 amino acids. In addition to these amino acids, our bodies contain several more, including selenocysteine (identical to cysteine but containing selenium rather than sulfur) and ornithine (remember this from glycolysis?). Amino acids can also undergo modifications: for instance, lysine residues can be acetylated. More amino acids and their variants are always being discovered as well.
Now that we have the building blocks of proteins established, the next blog post will focus on how these amino acids can be combined (polymerized) into long structures that make up polypeptides and proteins.
Brief aside: In my chemistry class in undergrad, my TA helped us remember the order of the chemical bonds following polymerization by saying N-H, C-H, C-O, N-H, C-H, C-O, ...
You may remember briefly from any stint in chemistry class that a carbon atom that is covalently bound to four different chemical entities (in this case, a side chain, a hydrogen atom, a carboxyl group, and an amino group) can take two different conformations, depending on how these bonds are spatially oriented. In the case of amino acids, the vast majority of amino acids found in our bodies and used to generate proteins are L stereoisomers. This is a result of the amino acid synthesis machinery structure exclusively generating L amino acids. There are exceptions, but we won't get into that.
As I mentioned, amino acids have a side chain: the part of the amino acid that endows it with its identity. These side chains can be broken into a few groups that we will explore now:
The first set of side chains is the nonpolar, hydrophobic side chains. The amino acids in this group include alanine, valine, leucine, isoleucine, glycine, methionine, and proline (structures shown to the right). What you'll immediately notice is that these amino acid side chains are composed mostly of hydrogens and carbons. Thus, these side chains do not contain polar covalent bonds and do not interact as readily with water (thus the term hydrophobic - they're "afraid" of water. Some amino acids of note in this group are proline, which contains a ring structure that creates a "kink" in the amino acid, and methionine, which contains a sulfur atom.
The next set is composed of the aromatic side chains, which includes phenylalanine, tyrosine, and tryptophan. These amino acids all contain an aromatic ring, which makes them relatively nonpolar; thus, they do not interact favorably with water. These amino acids are involved in mediating protein protein interactions and are frequently found at the active sites of enzymes.
Next up: polar, uncharged side chains: asparagine, cysteine, glutamine, serine, and threonine. These amino acids contain hydroxyl, sulfhydryl, or amide groups that mediate interactions with water, but they carry no net charge. An amino acid of note in this family is cysteine, which can react with itself to form cystine, which is important in mediating the formation of disulfide bonds in protein structures.
We'll consider basic side chains next. These amino acids consist of arginine, histidine, and lysine, which all carry a net positive charge in solution. Of note, histidine is commonly found at the active site of enzymes to serve as a protein donor or acceptor.
Finally, we find acidic side chains: aspartate and glutamate. In solution, these amino acids carry a negative charge and are considered acidic.
In the diagram at right, I've drawn up each of the amino acids along with their three-letter and one-letter codes. These codes are frequently used to abbreviate long lists of amino acids.
Another brief aside: Did you know that a single woman designated the amino acid abbreviations? She chose letters that made sense for most amino acids (as you can see above). For tryptophan, for example, she chose W because she envisioned saying tryptophan as twyptophan. Kind of cool, huh?
As a summary, here are the amino acid abbreviations:
- A, ala, alanine
- C, cys, cysteine
- D, asp, aspartate
- E, glu, glutamate
- F, phe, phenylalanine
- G, gly, glycine
- H, his, histidine
- I, ile, isoleucine
- K, lys, lysine
- L, leu, leucine
- M, met, methionine
- N, asn, asparagine
- P, pro, proline
- Q, gln, glutamine
- R, arg, arginine (think aRRRRginine)
- S, ser, serine
- T, thr, threonine
- V, val, valine
- W, trp, tryptophan (tWWWyptophan)
- Y, tyr, tyrosine
So there you have it: 20 amino acids. In addition to these amino acids, our bodies contain several more, including selenocysteine (identical to cysteine but containing selenium rather than sulfur) and ornithine (remember this from glycolysis?). Amino acids can also undergo modifications: for instance, lysine residues can be acetylated. More amino acids and their variants are always being discovered as well.
Now that we have the building blocks of proteins established, the next blog post will focus on how these amino acids can be combined (polymerized) into long structures that make up polypeptides and proteins.
Labels:
amino acids,
peptides,
polypeptide,
proteins
Friday, September 30, 2011
Android Applications for Scientists (and other people too!)
If you're like me, you like to use your phone for its capabilities, including for work, and because it's shiny and you paid a lot of money for it.
I use an Android phone (no thank you, Apple) and am always interested in an application that could help me with my research or studying. Unfortunately, the Android marketplace is cluttered with irrelevant applications, making finding useful applications difficult.
The following applications are presented as a summary of what I use. Certainly there are more out there (please tell me!) and more are created every day, and I look forward to using these applications in the future:
Astrid Tasks: Every morning as I'm walking into the building, I fill out my task list of things to do for the day. I don't need anything fancy - I just need an application that is quick, easy, and simple. Astrid is fantastic for putting together this list and for prioritizing my experiments and work. Plus, it's got a handy widget (if you're into widgets, that is). I would highly recommend this application for those who like to keep lists.
GoogleDocs: If you use Google for composing, sharing, or viewing documents, this application is fantastic because it will sync your computer and your phone to view the same documents. Additionally, if you've got a big enough screen, it's not too horrible to actually compose in these documents. I don't use this too often, but it's convenient when I've got a document to get on my phone and my Dropbox happens to be full (see below).
Doodle: Have you ever tried to schedule a meeting with faculty? How about with multiple faculty? Needless to say, it is a nightmare: herding cats as many would say. Doodle attempts to make this a little bit easier by creating a spreadsheet which participants can then check off for their ability. That one magical time spot that everyone checks is then the meeting time. Doodle comes with several options for creating an event and then adding participants. In my department, these things are really the best way to make sure that a meeting is really going to happen.
Dropbox: If you're not already on Dropbox, seriously, sign up for it. This program is the most useful thing I have every used. Once installed on a few computers and on your phone, a folder is created - your Dropbox. It acts just like a normal system folder and can be manipulated just like one too. The great thing is that anything that is in your Dropbox can be accessed on any computer that has Dropbox installed and you can also access your files online. This is so much better than carrying around a flash drive, and the syncing is instantaneous. I could not recommend this program more.
handyCalc: Usually, I'll use my crappy old calculator from high school to do the simple arithmetic needed for my experiments and notebook. When I'm at my computer and can't find my calculator, I use handyCalc, mainly because I find it much easier to use than the standard calculator that came with my phone. There are multiple iterations of calculators out there - from simple to mind-bogglingly complex. This program will solve equations, create graphs, and perform simple addition and subtraction. It works and I like it.
LinkedIn: At a recent conference, I learned the importance of LinkedIn. The seminar speaker asked everyone in the room to raise their hand if they were a member of LinkedIn. I was the only person not to raise my hand. LinkedIn is a Facebook for professionals - it can be helpful if you're looking for a job or want to make some contacts to look for a job in the future. Being able to use it on my phone is convenient, too.
Pulse: I like to read the news on my phone when I can't access a computer. The best app I've found for this is the Pulse reader app. Using this program, you can view tiles containing the top headlines from various websites, including numerous science-slanting websites. The handy swipe-to-change-story feature is nice, and the entire interface is easy-to-use. I also like using this while I'm walking because it's easy to pull up a short story that I can finish quickly and make myself feel like I accomplished something.
QuickOffice: I'm not a big fan of paid applications - I am a poor graduate student after all. QuickOffice came preinstalled with my phone, and I must say that I am quite impressed with it. It has the same features as the Microsoft Office suite (I can even view my PowerPoint slides on my phone!) and is really easy to use. If you're not up for shelling out $15 (!), you can always opt for the Google Docs app, which I think is almost just as good, and you can't beat the price...
Google Reader: I use Google Reader religiously for keeping up on journal articles. This handy little app presents my RSS feed conveniently and in my pocket so that I can keep up on my papers that I need to read. Since I can't access the actual papers on my phone, I star the articles of interest and download on my computer when I'm connected to my university's network.
The Weather Channel: I work in a lab with no windows. Most of the time, I can't tell is there's a tornado outside or a beautiful sunny day. Sometimes, that's for the best because then I'm not tempted to leave my benchtop and wander outside. When I do have to go outside, however, it's nice to know the weather, and the Weather Channel app is convenient for quickly checking the weather. Tons of functionality are included, such as animated weather maps. There are a lot of weather applications out there, but I find this to be the most useful.
WTFSIMFD: Hands down, my favorite food app. WTFSIMFD provides you with a recipes that you should eat. The app has a potty mouth, but it's amusing and endearing at the same time. It also gives great recipes, you know, for when you're not in the lab or studying...
Just a roundup of what I find useful as a student and a scientist. Maybe a followup post will be necessary when I find even more useful apps...
I use an Android phone (no thank you, Apple) and am always interested in an application that could help me with my research or studying. Unfortunately, the Android marketplace is cluttered with irrelevant applications, making finding useful applications difficult.
The following applications are presented as a summary of what I use. Certainly there are more out there (please tell me!) and more are created every day, and I look forward to using these applications in the future:
Astrid Tasks: Every morning as I'm walking into the building, I fill out my task list of things to do for the day. I don't need anything fancy - I just need an application that is quick, easy, and simple. Astrid is fantastic for putting together this list and for prioritizing my experiments and work. Plus, it's got a handy widget (if you're into widgets, that is). I would highly recommend this application for those who like to keep lists.
GoogleDocs: If you use Google for composing, sharing, or viewing documents, this application is fantastic because it will sync your computer and your phone to view the same documents. Additionally, if you've got a big enough screen, it's not too horrible to actually compose in these documents. I don't use this too often, but it's convenient when I've got a document to get on my phone and my Dropbox happens to be full (see below).
Doodle: Have you ever tried to schedule a meeting with faculty? How about with multiple faculty? Needless to say, it is a nightmare: herding cats as many would say. Doodle attempts to make this a little bit easier by creating a spreadsheet which participants can then check off for their ability. That one magical time spot that everyone checks is then the meeting time. Doodle comes with several options for creating an event and then adding participants. In my department, these things are really the best way to make sure that a meeting is really going to happen.
Dropbox: If you're not already on Dropbox, seriously, sign up for it. This program is the most useful thing I have every used. Once installed on a few computers and on your phone, a folder is created - your Dropbox. It acts just like a normal system folder and can be manipulated just like one too. The great thing is that anything that is in your Dropbox can be accessed on any computer that has Dropbox installed and you can also access your files online. This is so much better than carrying around a flash drive, and the syncing is instantaneous. I could not recommend this program more.
handyCalc: Usually, I'll use my crappy old calculator from high school to do the simple arithmetic needed for my experiments and notebook. When I'm at my computer and can't find my calculator, I use handyCalc, mainly because I find it much easier to use than the standard calculator that came with my phone. There are multiple iterations of calculators out there - from simple to mind-bogglingly complex. This program will solve equations, create graphs, and perform simple addition and subtraction. It works and I like it.
LinkedIn: At a recent conference, I learned the importance of LinkedIn. The seminar speaker asked everyone in the room to raise their hand if they were a member of LinkedIn. I was the only person not to raise my hand. LinkedIn is a Facebook for professionals - it can be helpful if you're looking for a job or want to make some contacts to look for a job in the future. Being able to use it on my phone is convenient, too.
Pulse: I like to read the news on my phone when I can't access a computer. The best app I've found for this is the Pulse reader app. Using this program, you can view tiles containing the top headlines from various websites, including numerous science-slanting websites. The handy swipe-to-change-story feature is nice, and the entire interface is easy-to-use. I also like using this while I'm walking because it's easy to pull up a short story that I can finish quickly and make myself feel like I accomplished something.
QuickOffice: I'm not a big fan of paid applications - I am a poor graduate student after all. QuickOffice came preinstalled with my phone, and I must say that I am quite impressed with it. It has the same features as the Microsoft Office suite (I can even view my PowerPoint slides on my phone!) and is really easy to use. If you're not up for shelling out $15 (!), you can always opt for the Google Docs app, which I think is almost just as good, and you can't beat the price...
Google Reader: I use Google Reader religiously for keeping up on journal articles. This handy little app presents my RSS feed conveniently and in my pocket so that I can keep up on my papers that I need to read. Since I can't access the actual papers on my phone, I star the articles of interest and download on my computer when I'm connected to my university's network.
The Weather Channel: I work in a lab with no windows. Most of the time, I can't tell is there's a tornado outside or a beautiful sunny day. Sometimes, that's for the best because then I'm not tempted to leave my benchtop and wander outside. When I do have to go outside, however, it's nice to know the weather, and the Weather Channel app is convenient for quickly checking the weather. Tons of functionality are included, such as animated weather maps. There are a lot of weather applications out there, but I find this to be the most useful.
WTFSIMFD: Hands down, my favorite food app. WTFSIMFD provides you with a recipes that you should eat. The app has a potty mouth, but it's amusing and endearing at the same time. It also gives great recipes, you know, for when you're not in the lab or studying...
Just a roundup of what I find useful as a student and a scientist. Maybe a followup post will be necessary when I find even more useful apps...
Saturday, September 24, 2011
Mendelian Genetics Part I
Gregor Mendel was an accomplished scientist, though he never would know this during his time. His studies of pea plants (as well as other organisms) laid the groundwork for the genetic breakthroughs that would come after his death. Mendel was a Augustinian monk that had ample time to experiment with his pea plants. The principles he was able to extract from his studies are the basics taught in high schools around the world.
Mendel studied peas, but not just any peas. His peas were true-breeding, meaning that they self-fertilized and produced essentially clones of themselves. Over the years of study, he bred plants with specific characteristics and then crossed different varieties of pea plants to test what their offspring would look like.
Importantly, Mendel considered discontinuous, contrasting traits: he only considered the traits that he could observe (such as color), came in a limited number of forms (green or yellow) and were easily distinguishable. For these monohybrid crosses, Mendel considered seven different traits: seed shape (round versus wrinkled), seed color (yellow versus green), pod shape (full versus constricted), pod color (yellow versus green), flower color (violet versus white), flower position (axial versus terminal), and stem height (tall versus short). Also important to Mendel's work was his use of mathematics and statistics to estimate the probabilities of a certain type of plant emerging from a certain type of pea plant cross.
The monohybrid cross is the consideration of one particular trait at a time. For example, if looking at pea pod color, one might cross a yellow and a green pea plant and then determine how many of the offspring had yellow or green pea pods. This type of cross is illustrated to the right in what's called a Punnett Square. A Punnett square is a way of organizing the different traits expressed by the individuals being crossed. The capitalized letters are the dominant traits, while the lower-cased letters are recessive. Vertical columns traditionally represent females; horizontal, males. Using the Punnett square, we can look at all the possible offspring that can emerge from a cross, which can then be used to determine probabilities associated with the offspring's traits.
From these monohybrid crosses, Mendel was able to make a few conclusions. When he crossed two pea plants of distinct traits (the P1 generation, true-breeding variety) to produce progeny (the F1 generation) and then used the progeny to generate more progeny (the F2 generation), he found that the two parental traits were still present and unchanged. This led to the hypothesis that each parent contributed equally to the inheritance of the "genetic determinants," which would have more technical and molecular definitions in the distant future. These determinants were separated and segregated randomly to make gametes and produce the next generation of pea plants.
Mendel's principles from the monohybrid crosses can be summarized as follows:
Mendel studied peas, but not just any peas. His peas were true-breeding, meaning that they self-fertilized and produced essentially clones of themselves. Over the years of study, he bred plants with specific characteristics and then crossed different varieties of pea plants to test what their offspring would look like.
Importantly, Mendel considered discontinuous, contrasting traits: he only considered the traits that he could observe (such as color), came in a limited number of forms (green or yellow) and were easily distinguishable. For these monohybrid crosses, Mendel considered seven different traits: seed shape (round versus wrinkled), seed color (yellow versus green), pod shape (full versus constricted), pod color (yellow versus green), flower color (violet versus white), flower position (axial versus terminal), and stem height (tall versus short). Also important to Mendel's work was his use of mathematics and statistics to estimate the probabilities of a certain type of plant emerging from a certain type of pea plant cross.
The monohybrid cross is the consideration of one particular trait at a time. For example, if looking at pea pod color, one might cross a yellow and a green pea plant and then determine how many of the offspring had yellow or green pea pods. This type of cross is illustrated to the right in what's called a Punnett Square. A Punnett square is a way of organizing the different traits expressed by the individuals being crossed. The capitalized letters are the dominant traits, while the lower-cased letters are recessive. Vertical columns traditionally represent females; horizontal, males. Using the Punnett square, we can look at all the possible offspring that can emerge from a cross, which can then be used to determine probabilities associated with the offspring's traits.
From these monohybrid crosses, Mendel was able to make a few conclusions. When he crossed two pea plants of distinct traits (the P1 generation, true-breeding variety) to produce progeny (the F1 generation) and then used the progeny to generate more progeny (the F2 generation), he found that the two parental traits were still present and unchanged. This led to the hypothesis that each parent contributed equally to the inheritance of the "genetic determinants," which would have more technical and molecular definitions in the distant future. These determinants were separated and segregated randomly to make gametes and produce the next generation of pea plants.
Mendel's principles from the monohybrid crosses can be summarized as follows:
- Hereditary determinants (unit factors) control traits that are in pairs in an individual.
- When two dissimilar unit factors for a trait are combined in one organism, one factor is dominant to the other recessive factor.
- When gametes are formed, the pair of unit factors separate and are equally likely to be separated into a gamete: the principle of segregation
In the next post, we'll examine dihybrid crosses and some more of the interesting findings from, of all things, pea plants.
Thursday, September 22, 2011
MAP Kinase Signaling
Cells exist in a very dynamic environment, not only on the inside of the cell membrane, but also on the outside. Thus, cells must be able to interact with their outside environment and respond to stimuli appropriately. One of the several signaling pathways involved in communication from the outside of the cell inward is the (very general) MAP kinase (MAPK) signaling pathway. The cell uses the MAPK signaling pathway for several reasons, primarily to amplify signals. Additionally, aberrant MAPK signaling is implicated in several types of cancers.
First, MAPK signifies mitogen activated protein kinase - a fancy word for a protein that responds to mitogens, or a molecule that stimulates mitosis. Proteins of the MAPK family were discovered in 1989 in yeast, with ERK1 being the first mammalian MAPK signaling protein, involved in insulin signaling pathways (at the time at least...).
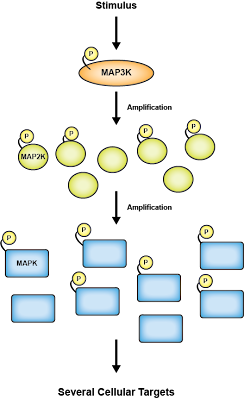
The MAPK signaling pathway is often called a cascade because the components in the pathway amplify a signal within the cell. The three major components are:
First, MAPK signifies mitogen activated protein kinase - a fancy word for a protein that responds to mitogens, or a molecule that stimulates mitosis. Proteins of the MAPK family were discovered in 1989 in yeast, with ERK1 being the first mammalian MAPK signaling protein, involved in insulin signaling pathways (at the time at least...).
The MAPK signaling pathway is often called a cascade because the components in the pathway amplify a signal within the cell. The three major components are:
- MAPKKK / MEKK / MAP3K: The MAP kinase kinase kinase. This protein phosphorylates MAP kinase kinase. Several forms are found within the cell and their abundance is lower than that of MAPKK or MAPK; thus, MAPKKK isn't as involved in the amplification of signals.
- MAPKK / MEK / MAP2K: The MAP kinase kinase. MAPKK phosphorylates MAPK and is highly abundant within the cell, but its only substrate is MAPK. Just a few phosphorylation events on MAPKK result in significant activation of MAPK.
- MAPK / ERK: The MAP kinase. MAPK is also abundant in the cell and has diverse substrates, including itself.
There are several pathways that involve MAPK signaling. Two of the important ones are:
- The ERK pathway: growth factor stimulation of cell surface receptors (via receptor tyrosine kinases [RTKs]) causes activation of Ras, which activates Raf (MAPKKK) to activate MEK (MAPKK) and then ERK (MAPK). ERK's several targets include Elk and Ets, which control cellular proliferation.
- The JNK pathway: Stress on the cell results in activation of several proteins, including Rho, which goes on to activate MEKK, then MEK, and JNK. JNK's major targets include c-Jun, ATF2, and Elk1, which control proliferation and apoptosis.
One important consideration is the specificity of MAPK signaling. Specificity can be achieved in several manners: protein components can only "fit" other specific protein components. Additionally, the spacial organization of a cell can affect specificity, and this type of specificity is often controlled by scaffold proteins.
MAPK signaling is essential for many cellular processes, including proliferation, apoptosis, embryonic development, and cancer progression. Many intricacies of the pathways are still being worked out and will provide significant insight in the future.
Thursday, September 1, 2011
The Basics of Protein Translation
Proteins, a major constituent of the cell, have many diverse functions and are classically considered to be the "work horses" of the cell. Additional molecules, such as RNAs and lipids, have shown importance in signaling and catalyzing chemical reactions; however, proteins remain an important part in the life of a cell.
Before proteins can perform their evolved functions, they must be synthesized within the cell. The process of synthesizing a protein is critically important to the cell and, thus, is an energy-intensive process.
The basic building blocks of a protein are amino acids, which come in twenty (and more) flavors. These amino acids have different properties that afford proteins different structures and functions when the amino acids are polymerized together in distinct orders. These amino acids can have distinct signaling roles when they exist as monomers as well (see this paper by Nobukuni et al for an example).
Monomeric amino acids in the cellular environment do not randomly polymerize to form proteins. In the first of several steps, tRNAs are charged: they are covalently linked to amino acids via aminoacyl tRNA synthetases. These synthetases hold the very important role of attaching the appropriate amino acid to the appropriate tRNA. Because inappropriate charging of tRNAs would lead to misincorporation of amino acids into a protein chain (wasting energy or leading to even bigger problems for the cell), sythetases are very specific. In fact, some synthetases have an editing site, where they will catalyze the removal of incorrectly placed amino acids.
After tRNAs are charged with their appropriate amino acids, they are ready for interaction with the ribosome. Ribosomes are large, complex molecules that merit their own post. Briefly, ribosomes are composed of RNA and protein and are made of two distinct complexes: the large and small subunits (depending on the origin of the ribosome, the subunits have different sedimentation coefficients, so you might see 30S and 50S for bacteria or 60S and 40S for eukaryotes, for example). Ribosomes catalyze the polymerization of amino acids into proteins.
In the first step of ribosome-mediated protein production, the small subunit of the ribosome combined with a tRNA for methionine (the amino acid that begins the protein chain) scans along the transcribed mRNA until it encounters a start site (ATG codon). Here, the complex stops, and the charged tRNA with its amino acid comes into contact with the peptidyl transferase site (P) on the ribosome. eIF2 (eukaryotic initiation factor 2), which was along for the ride, hydrolyzes GTP to GDP at this point such that the ribosome stops at the appropriate codon.
Next, the large subunit of the ribosome binds the small subunit, making a full ribosome that is ready for catalysis.
The tRNA that lines up with the mRNA's next codon then binds in the acceptor site (A), along with the help of eEF-1 (eukaryotic elongation factor-1), which hydrolyzes GTP to GDP. At this point, the ribosome goes into action: using the peptidyl transferase center (PTC), it catalyzes the covalent linkage of the first and second amino acids.
The entire ribosome now moves along the mRNA in a process called translocation, which requires the help of EF-2 (and GTP hydrolysis). The first tRNA is moved into the exit site (E), and the second tRNA moves into the P site, while the A site is open for another aminoacyl-tRNA.
The process repeats until the ribosome encounters a stop codon. At this point, termination factors (TFs), which have structures similar to tRNAs and bind mRNAs but do not have amino acids, enter the acceptor site of the ribosome. The ribosome then catalyzes the addition of water to the end of the amino acid chain, releasing it from the peptidyl transferase center and allowing it to leave the ribosome and begin folding into its native conformation.
While there are several details that I may have seemingly glazed over, this post should give a broad, simplified overview of translation. Future posts will address the many details involved.
In the first step of ribosome-mediated protein production, the small subunit of the ribosome combined with a tRNA for methionine (the amino acid that begins the protein chain) scans along the transcribed mRNA until it encounters a start site (ATG codon). Here, the complex stops, and the charged tRNA with its amino acid comes into contact with the peptidyl transferase site (P) on the ribosome. eIF2 (eukaryotic initiation factor 2), which was along for the ride, hydrolyzes GTP to GDP at this point such that the ribosome stops at the appropriate codon.
Next, the large subunit of the ribosome binds the small subunit, making a full ribosome that is ready for catalysis.
The tRNA that lines up with the mRNA's next codon then binds in the acceptor site (A), along with the help of eEF-1 (eukaryotic elongation factor-1), which hydrolyzes GTP to GDP. At this point, the ribosome goes into action: using the peptidyl transferase center (PTC), it catalyzes the covalent linkage of the first and second amino acids.
The entire ribosome now moves along the mRNA in a process called translocation, which requires the help of EF-2 (and GTP hydrolysis). The first tRNA is moved into the exit site (E), and the second tRNA moves into the P site, while the A site is open for another aminoacyl-tRNA.
The process repeats until the ribosome encounters a stop codon. At this point, termination factors (TFs), which have structures similar to tRNAs and bind mRNAs but do not have amino acids, enter the acceptor site of the ribosome. The ribosome then catalyzes the addition of water to the end of the amino acid chain, releasing it from the peptidyl transferase center and allowing it to leave the ribosome and begin folding into its native conformation.
While there are several details that I may have seemingly glazed over, this post should give a broad, simplified overview of translation. Future posts will address the many details involved.
Labels:
amino acids,
polypeptide,
protein synthesis,
ribosome,
tRNA
Saturday, August 27, 2011
The Many Functions of Ubiquitin
Ubiquitin (abbreviated as Ub) is described as a small, frequently-encountered molecule involved in the proteosomal degradation of proteins. In reality, the addition of ubiquitin moieties to proteins can serve several functions and is now recognized as a common post-translational modification (PTM).
The classical ubiquitin pathway that first comes to mind is that of proteosomal degradation. In this pathway, illustrated in the accompanying figure, ubiquitin is first "activated" by binding covalently to a cysteine residue on E1, the ubiquitin-activating enzyme, which requires the energy of ATP. Then ubiquitin is transfered to a cysteine residue on E2, the ubiquitin-conjugating enzyme (no ATP is required here). Finally, E2 binds to a scaffold called E3, the ubiquitin ligase, which also contains the target protein - illustrated in pink. This target protein is then covalently linked to ubiquitin via a lysine residue. Several rounds of this pathway then result in linear polyubiquitination of proteins, which is then recognized by other components in the cell that result in its recruitment to the proteasome and subsequent degradation (a topic of future posts). The end result is that the target protein is broken down into small seven-to-eight amino acid pieces.
As mentioned above, there are three proteins involved in the attachment of ubiquitin to target proteins, E1, E2, and E3. In the cell, E1s are the least plentiful (only one has been discovered thus far), while the human genome codes for tens of E2. Several more E3 ligases have been discovered (and more every day).
Importantly, ubiquitination does not exclusively function in proteosome-mediated degradation of proteins. Monoubiquitination, or the attachment of a single ubiquitin moiety to a protein, has been shown to have diverse effects on target proteins (see listbelow). Additionally, multiple monoubiquitination events can occur on a single protein, as can branched polyubiquitination.
Ubiquitin is not the sole small protein modifier. Several molecules similar to ubiquitin have been described and exhibit diverse functions in cellular signaling (topics of future posts). These proteins include SUMO (small ubiquitin-like modifier), NEDD, FATIO, and FUBI. Needless to say, all of these proteins have some of the most adorable names in science.
The following list is a short compilation of ubiquitin functions, and, as we all know very well, no hard-and-fast rules exist for ubiquitin modifications (or science in general).
Some cool (and important) papers and links:
Principles of ubiquitin and SUMO modifications in DNA repair
Non-traditional Functions of Ubiquitin and Ubiquitin-binding Proteins
The Ubiquitin System
Nonproteolytic Functions of Ubiquitin in Cell Signaling
Ubiquitin Function and Variety
The classical ubiquitin pathway that first comes to mind is that of proteosomal degradation. In this pathway, illustrated in the accompanying figure, ubiquitin is first "activated" by binding covalently to a cysteine residue on E1, the ubiquitin-activating enzyme, which requires the energy of ATP. Then ubiquitin is transfered to a cysteine residue on E2, the ubiquitin-conjugating enzyme (no ATP is required here). Finally, E2 binds to a scaffold called E3, the ubiquitin ligase, which also contains the target protein - illustrated in pink. This target protein is then covalently linked to ubiquitin via a lysine residue. Several rounds of this pathway then result in linear polyubiquitination of proteins, which is then recognized by other components in the cell that result in its recruitment to the proteasome and subsequent degradation (a topic of future posts). The end result is that the target protein is broken down into small seven-to-eight amino acid pieces.
As mentioned above, there are three proteins involved in the attachment of ubiquitin to target proteins, E1, E2, and E3. In the cell, E1s are the least plentiful (only one has been discovered thus far), while the human genome codes for tens of E2. Several more E3 ligases have been discovered (and more every day).
Importantly, ubiquitination does not exclusively function in proteosome-mediated degradation of proteins. Monoubiquitination, or the attachment of a single ubiquitin moiety to a protein, has been shown to have diverse effects on target proteins (see listbelow). Additionally, multiple monoubiquitination events can occur on a single protein, as can branched polyubiquitination.
Ubiquitin is not the sole small protein modifier. Several molecules similar to ubiquitin have been described and exhibit diverse functions in cellular signaling (topics of future posts). These proteins include SUMO (small ubiquitin-like modifier), NEDD, FATIO, and FUBI. Needless to say, all of these proteins have some of the most adorable names in science.
The following list is a short compilation of ubiquitin functions, and, as we all know very well, no hard-and-fast rules exist for ubiquitin modifications (or science in general).
- p53 ubiquitination by MDM2: cell cycle regulation, apoptosis
- HIF1 ubiquitination by VHL: hypoxic response
- Caspase 3 and 7 ubiquitination by XIAP: apoptosis
- PCNA: DNA repair
- RNA polymerase II: transcriptional regulation
- H2AX and H2A: transcriptional regulation, protein recruitment
Of course, there are more proteins that are modified by ubiquitin, but I hope that the above table illustrates the diversity of the modification.
Interestingly, ubiquitin modifications are not forever: de-ubiquitinating enzymes (DUBs) remove ubiquitin moieties from tagged proteins. Thus, the reversible ubiquination represents a dynamic cellular process.
Some cool (and important) papers and links:
Principles of ubiquitin and SUMO modifications in DNA repair
Non-traditional Functions of Ubiquitin and Ubiquitin-binding Proteins
The Ubiquitin System
Nonproteolytic Functions of Ubiquitin in Cell Signaling
Ubiquitin Function and Variety
Subscribe to:
Posts (Atom)