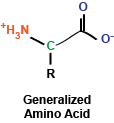The last post about an operon (the lac operon) is the most viewed post on this blog, so I thought that it might be helpful to follow this up with another operon, this time concentrating on the trp (tryptophan) operon. This operon is another really elegant example of transcriptional regulation in E.coli and the mechanism is pretty cool.
Amino acids are essential for life (see the last post on their composition!) and cells synthesize amino acids using a variety of enzymes. When nutrients are plentiful, such as E.coli would encounter in nutrient broth in the laboratory setting, cells no longer need to waste energy producing biosynthetic enzymes when they can utilize nutrients already in excess. The trp operon contains several enzymes that are coordinately regulated and involved in the production of tryptophan. When tryptophan is present in the cell's environment, it doesn't need to make any of these enzymes, but if the cell needs tryptophan, these enzymes are transcribed and shortly thereafter translated. Control of this operon, thus, controls how much energy the cell is going to put into making tryptophan.
Similar to the lac operon, the trp operon contains an operator (O) sequence, within the promoter sequence, where an operator binds and prevents transcription. In the presence of tryptophan, the operator binds the promoter and prevents RNA polymerase from transcribing genes. In the absence of tryptophan, however, transcription occurs at a basal rate. Sounds simple enough, right? Let's take it a step further and consider...
Attenuation
An important concept in gene regulation is that of attenuation, which is fine-tuning of gene expression. You might think that attenuation is mediated by protein factors that bind the DNA and affect gene expression; however, attenuation of the trp operon is a little different and, instead, depends on mRNA structure to modulate gene expression.
Before moving forward, let's look at the trp operon (diagrammed to the right). Briefly,t here are four regions, and these four regions have differing levels of complementarity to each other. Thus, when the DNA is transcribed into mRNA, the mRNA folds into all kinds of shapes and the regions of the trp operon fold on each other.
After transcription of the entire trp operon (we're dealing with mRNA from this point forward), the next event is translation of this mRNA into protein. In bacteria, it's important to note that transcription and translation occur simultaneously, so as soon as we have a transcript in a bacterial cell, it's being translated. The trp transcript contains two critical tryptophan codons immediately before region 1, so in order to synthesize the enzymatic machinery to make tryptophan, the cell must use a few residues to translate the protein.
In the presence of high amounts of tryptophan within the cell, the ribosome plows through these two tryptophan codons, adding in the appropriate amino acids, and continuing through region 1 of the mRNA. This results in region 1 and 2 mRNA sequences binding together, and then regions 3 and 4 bind together as well. This interaction between regions 3 and 4 results in the creation of a transcription-termination hairpin, basically a structure in the mRNA that kicks out RNA polymerase and prevents further transcription of the mRNA. Thus, transcription (and then translation) are stopped because
In the absence of tryptophan, however, the ribosome cannot quickly add tryptophan during the translation process and it stalls before region 1. This results in the folding of the mRNA such that regions 2 and 3 bind to each other. When this structure forms, no transcriptional termination hairpin is formed, and mRNA synthesis continues. Thus, the entire mRNA sequence for the trp operon is made and can be translated into enzymes that will synthesize tryptophan.
In summary:
Lots of tryptophan: Ribosome zooms through the mRNA, regions 1 & 2 and 3 & 4 bind (in pairs) and create a termination hairpin
End result: Transcription terminates and tryptophan synthetic enzymes not created (cell saves energy!)
Lack of tryptophan: Ribosome stalls immediately before region 1, regions 2 and 3 bind each other, no termination hairpin is formed
End result: Transcription continues and biosynthetic enzymes are eventually synthesized
This scheme is similar for other operons encoding amino acid biosynthetic enzymes (in bacteria, that is). The trp operon is an elegant scheme to finely-tune transcription via mRNA structure to prevent the cell from wasting energy.
Monday, October 17, 2011
The Trp Operon
Labels:
amino acids,
gene regulation,
lac operon,
operator,
operon,
transcription,
trp,
tryptophan
Thursday, October 13, 2011
Amino Acids: The Building Blocks of Proteins
While I've written many posts describing signaling pathways and cellular phenomena of significant complexity, I'd like to use this post to take a step back and look at some fundamental building blocks, first turning to amino acids, the monomers that, when polymerized, make up polypeptides and proteins.
At the most basic level, amino acids are really a rather simple chemical compound, consisting of a amino group, a carboxy group, a hydrogen, and a side chain, all sticking off a central carbon atom. These amino acids are polymerized via their amino and carboxy chemical groups to create long, linear linkages.
Brief aside: In my chemistry class in undergrad, my TA helped us remember the order of the chemical bonds following polymerization by saying N-H, C-H, C-O, N-H, C-H, C-O, ...
You may remember briefly from any stint in chemistry class that a carbon atom that is covalently bound to four different chemical entities (in this case, a side chain, a hydrogen atom, a carboxyl group, and an amino group) can take two different conformations, depending on how these bonds are spatially oriented. In the case of amino acids, the vast majority of amino acids found in our bodies and used to generate proteins are L stereoisomers. This is a result of the amino acid synthesis machinery structure exclusively generating L amino acids. There are exceptions, but we won't get into that.
As I mentioned, amino acids have a side chain: the part of the amino acid that endows it with its identity. These side chains can be broken into a few groups that we will explore now:
The first set of side chains is the nonpolar, hydrophobic side chains. The amino acids in this group include alanine, valine, leucine, isoleucine, glycine, methionine, and proline (structures shown to the right). What you'll immediately notice is that these amino acid side chains are composed mostly of hydrogens and carbons. Thus, these side chains do not contain polar covalent bonds and do not interact as readily with water (thus the term hydrophobic - they're "afraid" of water. Some amino acids of note in this group are proline, which contains a ring structure that creates a "kink" in the amino acid, and methionine, which contains a sulfur atom.
The next set is composed of the aromatic side chains, which includes phenylalanine, tyrosine, and tryptophan. These amino acids all contain an aromatic ring, which makes them relatively nonpolar; thus, they do not interact favorably with water. These amino acids are involved in mediating protein protein interactions and are frequently found at the active sites of enzymes.
Next up: polar, uncharged side chains: asparagine, cysteine, glutamine, serine, and threonine. These amino acids contain hydroxyl, sulfhydryl, or amide groups that mediate interactions with water, but they carry no net charge. An amino acid of note in this family is cysteine, which can react with itself to form cystine, which is important in mediating the formation of disulfide bonds in protein structures.
We'll consider basic side chains next. These amino acids consist of arginine, histidine, and lysine, which all carry a net positive charge in solution. Of note, histidine is commonly found at the active site of enzymes to serve as a protein donor or acceptor.
Finally, we find acidic side chains: aspartate and glutamate. In solution, these amino acids carry a negative charge and are considered acidic.
In the diagram at right, I've drawn up each of the amino acids along with their three-letter and one-letter codes. These codes are frequently used to abbreviate long lists of amino acids.
Another brief aside: Did you know that a single woman designated the amino acid abbreviations? She chose letters that made sense for most amino acids (as you can see above). For tryptophan, for example, she chose W because she envisioned saying tryptophan as twyptophan. Kind of cool, huh?
As a summary, here are the amino acid abbreviations:
So there you have it: 20 amino acids. In addition to these amino acids, our bodies contain several more, including selenocysteine (identical to cysteine but containing selenium rather than sulfur) and ornithine (remember this from glycolysis?). Amino acids can also undergo modifications: for instance, lysine residues can be acetylated. More amino acids and their variants are always being discovered as well.
Now that we have the building blocks of proteins established, the next blog post will focus on how these amino acids can be combined (polymerized) into long structures that make up polypeptides and proteins.
Brief aside: In my chemistry class in undergrad, my TA helped us remember the order of the chemical bonds following polymerization by saying N-H, C-H, C-O, N-H, C-H, C-O, ...
You may remember briefly from any stint in chemistry class that a carbon atom that is covalently bound to four different chemical entities (in this case, a side chain, a hydrogen atom, a carboxyl group, and an amino group) can take two different conformations, depending on how these bonds are spatially oriented. In the case of amino acids, the vast majority of amino acids found in our bodies and used to generate proteins are L stereoisomers. This is a result of the amino acid synthesis machinery structure exclusively generating L amino acids. There are exceptions, but we won't get into that.
As I mentioned, amino acids have a side chain: the part of the amino acid that endows it with its identity. These side chains can be broken into a few groups that we will explore now:
The first set of side chains is the nonpolar, hydrophobic side chains. The amino acids in this group include alanine, valine, leucine, isoleucine, glycine, methionine, and proline (structures shown to the right). What you'll immediately notice is that these amino acid side chains are composed mostly of hydrogens and carbons. Thus, these side chains do not contain polar covalent bonds and do not interact as readily with water (thus the term hydrophobic - they're "afraid" of water. Some amino acids of note in this group are proline, which contains a ring structure that creates a "kink" in the amino acid, and methionine, which contains a sulfur atom.
The next set is composed of the aromatic side chains, which includes phenylalanine, tyrosine, and tryptophan. These amino acids all contain an aromatic ring, which makes them relatively nonpolar; thus, they do not interact favorably with water. These amino acids are involved in mediating protein protein interactions and are frequently found at the active sites of enzymes.
Next up: polar, uncharged side chains: asparagine, cysteine, glutamine, serine, and threonine. These amino acids contain hydroxyl, sulfhydryl, or amide groups that mediate interactions with water, but they carry no net charge. An amino acid of note in this family is cysteine, which can react with itself to form cystine, which is important in mediating the formation of disulfide bonds in protein structures.
We'll consider basic side chains next. These amino acids consist of arginine, histidine, and lysine, which all carry a net positive charge in solution. Of note, histidine is commonly found at the active site of enzymes to serve as a protein donor or acceptor.
Finally, we find acidic side chains: aspartate and glutamate. In solution, these amino acids carry a negative charge and are considered acidic.
In the diagram at right, I've drawn up each of the amino acids along with their three-letter and one-letter codes. These codes are frequently used to abbreviate long lists of amino acids.
Another brief aside: Did you know that a single woman designated the amino acid abbreviations? She chose letters that made sense for most amino acids (as you can see above). For tryptophan, for example, she chose W because she envisioned saying tryptophan as twyptophan. Kind of cool, huh?
As a summary, here are the amino acid abbreviations:
- A, ala, alanine
- C, cys, cysteine
- D, asp, aspartate
- E, glu, glutamate
- F, phe, phenylalanine
- G, gly, glycine
- H, his, histidine
- I, ile, isoleucine
- K, lys, lysine
- L, leu, leucine
- M, met, methionine
- N, asn, asparagine
- P, pro, proline
- Q, gln, glutamine
- R, arg, arginine (think aRRRRginine)
- S, ser, serine
- T, thr, threonine
- V, val, valine
- W, trp, tryptophan (tWWWyptophan)
- Y, tyr, tyrosine
So there you have it: 20 amino acids. In addition to these amino acids, our bodies contain several more, including selenocysteine (identical to cysteine but containing selenium rather than sulfur) and ornithine (remember this from glycolysis?). Amino acids can also undergo modifications: for instance, lysine residues can be acetylated. More amino acids and their variants are always being discovered as well.
Now that we have the building blocks of proteins established, the next blog post will focus on how these amino acids can be combined (polymerized) into long structures that make up polypeptides and proteins.
Labels:
amino acids,
peptides,
polypeptide,
proteins
Subscribe to:
Posts (Atom)